One Way to Prevent Cancer: Map the Fundamentals of How Cells Go Awry
Thursday, October 8, 2020
Someday, scientists may be able to prevent cancer by controlling two proteins that operate deep inside the quagmire of epigenetic cell fate transitions, a new paper suggests.
Published by Nature Communications, the article describes a dynamic push and pull between the proteins ANP32E and H2AZ. Their relationship is significant because too little of H2AZ promotes cell division and aggressive tumors, while high levels of H2AZ promotes chaos and metastatic cancer — and ANP32E acts as a "chaperone" that directs H2AZ, said the study's corresponding author, Patrick Murphy, Ph.D. He is an assistant professor of Biomedical Genetics at the University of Rochester Medical Center and a Wilmot Cancer Institute investigator.
Abundant levels of H2AZ are often found in people with breast and brain cancer and melanoma.
With a "normal" amount of H2AZ, Murphy said, healthy cells can turn off the signals that permit cancer cells to divide and tumors to grow.
Epigenetics is at the root of Murphy's work. The field has been around for 80 years but is getting a lot of buzz more recently. Scientists and cancer patients are learning what biological factors influence inherited gene changes and predisposition to diseases, and how lifestyle behaviors passed down through generations and chemical exposures can also switch the function of genes and lead to cancer and other illnesses.
The core of epigenetics is understanding how two cells can have the same DNA but have different functions in the body. The work by the Murphy lab adds a critical bit of information as scientists clamor to find the factors that activate or silence genes during cell transitions.
Doug Portman Becomes New Director of the GDSC Graduate Program
Friday, September 18, 2020

We are delighted to announce that Dr. Portman will assume the directorship of the graduate program in Genetics, Development and Stem Cells. Both a geneticist and neuroscientist at heart, Doug has been a long-standing member of the Department of Biomedical Genetics with joint appointments in the Departments of Biology and Neuroscience. He has also served as the director of the admission committee for the neuroscience graduate program and has been a reviewer and panel chair for the prestigious Ruth L. Kirchstein National Research Service Award fellowship program. He brings an extensive experience in graduate education to the table and we are excited that he has agreed to lead our GDSC program forward! Congratulations Doug!
After Five Years at the Helm, Chris Pröschel Hands Off GDSC Program leadership
Thursday, September 17, 2020

After five successful years of service Chris Pröschel has elected to step down from the directorship of the Genetics, Development and Stem Cell graduate program. During his tenure he introduced new training opportunities, started outreach activities, initiated new student events and created a home for our first-year students. He also helped integrate the program faculty into an inclusive admission process which has resulted in the rapid growth of the program. "To me, it has been both an exhilarating and exhausting time" Chris said. "There are so many people that I want to thank for their help and support! In addition to our faculty and staff, this includes our friends at GEPA, all of whom are tireless advocates for graduate education. But most of all I thank the students - past and present - who are the heart and soul and of program!". Chris looks forward to spending more time on his very active research program, and remains an integral part of our community through his many collaborations with UR investigators.
GDSC Predoc Adrián Moisés Molina Vargas Passes his Qualifying Exam!
Thursday, September 3, 2020

Adrian Vargas, a pre-doctoral student in the laboratory of Dr. Mitch O'Connell successfully defended his Qualifying Exam proposal entitled "Understanding guide-RNA design rules for efficient and specific Cas13 RNA-binding in human cells". Working on the recently described Type VI CRISPR-Cas system Adrian is investigating unique properties of the effector nuclease Cas13. His work has designed new approaches that will shed light on the mechanisms that govern optimal Cas13 RNA-binding, and potentially enhance its usability in human cells. Pursuing such goals not only will help complete our understanding of the gRNA requirements for on-target efficiency but more importantly, it will assist in uncovering what gRNA features contribute to off-target effects that could hinder the use of Cas13 as a specific RNA-targeting tool. The results will develop a more holistic biophysical model of Cas13 binding and cleavage activity. Congratulations Adrian on a job well done! You can read Adrian's thesis abstract below.
Title: Understanding guide-RNA design rules for efficient and specific Cas13 RNA-binding in human cells.
Abstract: Prokaryotic adaptive immune systems utilize Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) and CRISPR Associated (Cas) proteins to target and cleave foreign genetic elements in an RNA-guided manner. The recently described Type VI CRISPR-Cas system contains a single effector ribonuclease, Cas13, that binds and processes a guide-RNA (gRNA), forming an RNA-guided RNA-targeting protein complex. Cas13 has been successfully engineered for potent RNA-knockdown in eukaryotic cells with minimal off-target effects, and other exciting applications described to date include utilizing a nuclease-dead Cas13 (dCas13) as a programmable RNA-binding protein for RNA imaging, RNA-splicing, RNA-detection or RNA-editing applications, among others.
However, the principles of guide-RNA selection for efficient and specific Cas13:gRNA RNA binding remain elusive. Previous work indicates that gRNA choice is important because not all gRNAs yield a robust RNA-targeting, and it is likely that there are different gRNA requirements for RNA-binding compared to RNA-cleavage applications. Furthermore, no predictive models exist for guide-RNA binding efficiency and specificity for Cas13 because no systematic screens have been carried out. Given that no transcriptome-wide measurements of Cas13-binding specificity have been attempted so far, it is unknown what the Cas13 off-target landscape is for human cells. Addressing this knowledge gap and quantitatively understanding the differences between binding and cleavage specificities is integral to the development of specific Cas13 RNA-binding applications.
In this project, I propose a series of approaches that will shed light on the mechanisms that govern optimal Cas13 RNA-binding, and potentially enhance its usability in human cells. Pursuing such goals not only will help complete our understanding of the gRNA requirements for on-target efficiency but more importantly, it will assist us in uncovering what gRNA features contribute to off-target effects that could hinder the use of Cas13 as a specific RNA-targeting tool, particularly for RNA-binding applications. In addition, these findings will be complemented by kinetic studies to dissect the biochemical components underlying Cas13 binding specificity to develop a more holistic biophysical model of Cas13 binding and cleavage activity. Consequently, we will generate a predictive model for Cas13 guide-RNA targeting specificity which will be complied into a comprehensive gRNA design tool tailored to different RNA-targeting applications for any RNA of choice. Finally, using the knowledge we generate and existing structural information, we will attempt to rationally engineer Cas13 and gRNA structure to enhance binding (and cleavage) specificities. Taken together, this work will bolster efforts to develop a range of RNA-targeting tools that can be readily used as research tools to address knowledge gaps in RNA biology, and as potential therapeutics.
GDSC Predoc Tom O’Connor Passes his Qualifying Exam!
Thursday, August 20, 2020

Having recently completed his second year, earlier this month, GDSC Predoc Tom O'Connor passed his Qualifying Exam. Tom is pursuing his Ph.D. research under a co-mentorship with Dr. Robert Dirksen and Dr. Joe Chakkalakal. His doctoral research project is titled "Endurance exercise as a therapeutic intervention to preserve skeletal muscle function post-radiation." His research abstract is sampled below. Congratulations Tom!
Abstract: Exercise has several beneficial effects on overall health, such as increased bone strength and improved cardiovascular, cognitive, and immune system function. Similarly, exercise elicits improved muscle function by increasing muscle stem cell (satellite cell, SC) activation and improving excitation-contraction coupling through the establishment of Ca2+ Entry Units (CEUs) and upregulation of calcium handling genes. However, radiotherapy, a mainstay in the treatment of many cancers, produces multiple adverse effects on skeletal muscle and has been shown to cause early-onset sarcopenia, a condition characterized by significant muscle weakening commonly observed in the elderly. Our studies show that radiation produces cytotoxic effects on SCs, depleting the SC pool and leaving the remaining SCs dysfunctional in their ability to activate and repair injured myofibers. We find that radiation exposure also decreases expression of key Ca2+ handling genes in skeletal muscle including CASQ1, CACNB1, and STIM1. Calsequestrin-1 binds Ca2+ in the sarcoplasmic reticulum (SR), buffering intracellular Ca2+ stores upon muscle excitation-contraction. CACNB1 encodes the b subunit of the 5 subunit dihydropyridine receptor (DHPR), the voltage sensor in the transverse tubule (TT) that activates ryanodine receptor (RyR1) in the SR to release Ca2+ for muscle contraction. STIM1 encodes the stromal-interaction molecule-1, a Ca2+ sensor in the SR, which activates Orai1 Ca2+-permeable channels in the TT when SR Ca2+ stores become depleted, a process known as store-operated calcium entry (SOCE).
The central hypothesis of this project is that post-radiation exercise restores muscle repair and regeneration function through activation of the remaining SC pool, allowing for turnover of damaged myonuclei. We also postulate that endurance exercise restores homeostatic Ca2+ control mechanisms after radiation through upregulation of integral Ca2+ handling genes and establishment of CEUs, overall restoring post-radiation muscle contractile function. I will assess the effect of post-radiation chronic endurance exercise on physiological muscle function using a juvenile radiation mouse model. Aim 1 will characterize SC adaptation in terms of myonuclear contribution and regenerative function post-voluntary wheel running (VWR), the model we will use for chronic endurance exercise. Aim 2 will address how Ca2+ handling is affected by VWR, assessing SOCE, gene expression, and resting calcium levels. Aim 3 will address the effects of radiation on SCs and Ca2+ handling and the therapeutic extent of post-radiation VWR to restore physiological muscle function. The results of these studies will provide novel insight into the patho-mechanisms of radiation-induced sarcopenia and determine the therapeutic potential of chronic endurance exercise to rescue SC regenerative capacity and enhance Ca2+ handling in an attempt to restore overall muscle function.
BMG Faculty, Douglas Portman, Ph.D. Leads Study: Biology Blurs Line Between Sexes, Behaviors
Monday, August 10, 2020
A new study, from the journal Current Biology, spotlights research from Biomedical Genetics Professor Douglas Portman, Ph.D. Titled Dynamic, Non-binary Specification of Sexual State in the C. elegans Nervous System, this research "identifies a genetic switch in brain cells that can toggle between sex-specific states when necessary, findings that question the idea of sex as a fixed property". Further information can be found on the URMC's Del Monte Institute's webpage: NeURoscience.
Xiaolu Wei awarded the University of Rochester Agnes and George Messersmith Dissertation Fellowship for 2020-2021!
Tuesday, June 9, 2020
 Congratulations to Xiaolu Wei in the GDSC Program on receiving this prestigious award! Xiaolu's work in the Larracuente Lab combines genomic, and molecular techniques to gain insight into how large blocks of repetitive DNA called satellites are regulated in the germline. Satellite DNAs are abundant in genomes but their functions are unknown because their repetitive nature makes them extremely difficult to study. The little that we know suggest that they have important roles in chromosome segregation and in genome evolution, and are often associated with human diseases and speciation. Xiaolu is taking a two-pronged approach to studying satellite DNA regulation. First, she is asking broad questions about how satellites are regulated at the transcriptional and chromatin level during gametogenesis. Second, she is studying the molecular mechanism of a selfish meiotic drive system in Drosophila melanogaster males called Segregation Distorter (SD). In this system, SD kills sperm that carry a particular satellite DNA, through a chromatin condensation defect. She aims to use this system to gain insight into satellite regulation in the male germline.
Congratulations to Xiaolu Wei in the GDSC Program on receiving this prestigious award! Xiaolu's work in the Larracuente Lab combines genomic, and molecular techniques to gain insight into how large blocks of repetitive DNA called satellites are regulated in the germline. Satellite DNAs are abundant in genomes but their functions are unknown because their repetitive nature makes them extremely difficult to study. The little that we know suggest that they have important roles in chromosome segregation and in genome evolution, and are often associated with human diseases and speciation. Xiaolu is taking a two-pronged approach to studying satellite DNA regulation. First, she is asking broad questions about how satellites are regulated at the transcriptional and chromatin level during gametogenesis. Second, she is studying the molecular mechanism of a selfish meiotic drive system in Drosophila melanogaster males called Segregation Distorter (SD). In this system, SD kills sperm that carry a particular satellite DNA, through a chromatin condensation defect. She aims to use this system to gain insight into satellite regulation in the male germline.
Researchers Find New Leukemia Genes using CRISPR Technology
Monday, April 20, 2020
Using the most advanced tools available, scientists discovered several novel genes not known to be involved in blood cancers, and used the powerful new data to paint a clearer map for how aggressive leukemia arises and grows, according to an article published in Nature Cancer.
Jeevisha Bajaj, Ph.D., assistant professor of Biomedical Genetics at the University of Rochester Medical Center and a researcher at the Wilmot Cancer Institute, is the lead author of the study. Bajaj conducted the research while she was a project scientist in the laboratory of Tannishtha Reya Ph.D., professor of Pharmacology and Medicine at the University of California, San Diego School of Medicine, and senior author of the study.
The paper points to several significant discoveries:
- It unveiled a new gene, Staufen 2 (Stau2), that regulates and drives the molecular programs for leukemia stem cells, the cells responsible for propagating the disease and for therapy resistance. Stau2 has been previously studied in the brain and nervous system but until now was not known to have a role in cancer.
- The team used a tool known as CRISPR, which allows scientists to edit DNA in cells and focus on large groups of genes active in a particular disease - in this case, myeloid leukemias. The paper showed that CRISPR can identify an entire class of gene mediators for leukemia, which will aid future research.
- The team also tested its hypothesis in a mouse model designed to mimic the human experience with leukemia, as opposed to conducting studies solely in cell cultures, as several other groups had previously done.
A Big THANK YOU to Our Graduate Student Researchers!
Thursday, April 9, 2020
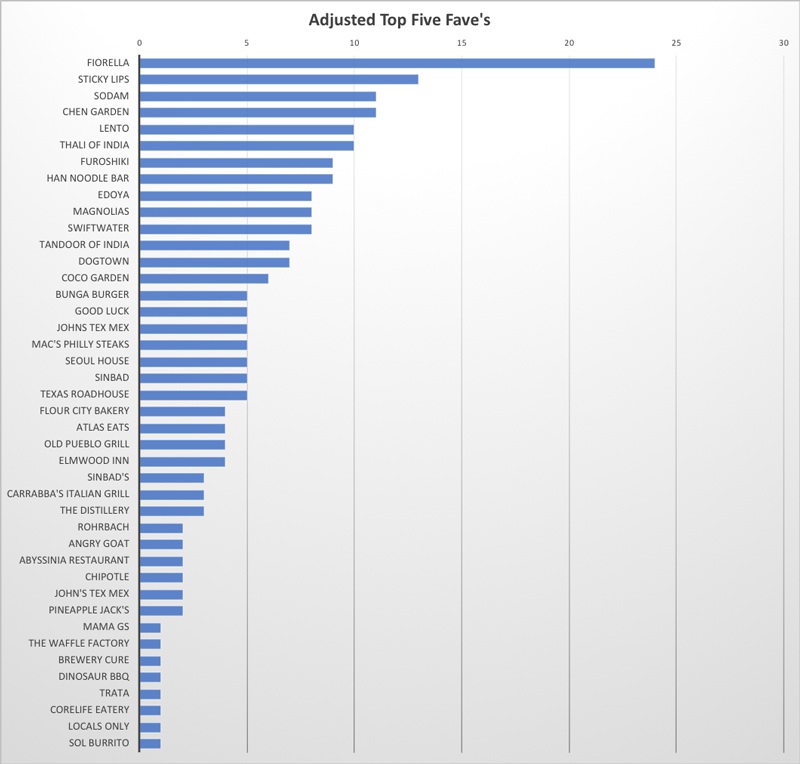
As our students carry-on their work sheltered away at home, there is much food for thought to be had. But there is also an opportunity to enjoy food from our community restaurants, which provides a much needed change in pace. So we asked our GDSC program students to rank their five favorite restaurants. The results were weighted based on ranking (#1-#5), and are tabulated in the graph. Clearly we are fortunate to have a wide variety of restaurants at our finger-tips! And for many there will be new ones to try based on the list below. As one faculty put it "Wow, I only know four restaurants from this list". Clearly -- here is an opportunity to switch up our lives and also support our culinary neighbors.
This survey was part of our Graduate Student Appreciation Week, which we celebrated appropriately with restaurant gift cards from the mentors and the GDSC program to all our hard-working graduate student researchers! Thank you --Stay smart and stay well!